Transcription: A hub of activity
It is exceedingly rare that a protein acts alone – more often, proteins co-operate with one another so that they can function with greater speed, specificity, or reactivity. The process by which they assemble at promoter regions within the genome in order to initiate gene transcription is relatively well understood. However, much less is known about how proteins come together at enhancer regions – sites that regulate gene expression – and how each protein contributes to transcription.
One form of co-operation involves transcription factors and other regulatory proteins physically associating with one another to form a ‘hub’ – a pocket of high protein concentration – around gene enhancers and promoters in the cell nucleus (Hnisz et al., 2017). But how do these hubs form and evolve over time? How do they ‘sense’ upstream signaling input? And how do they foster transcription? Now, in eLife, Sarah Bray from the University of Cambridge and colleagues – including Javier deHaro-Arbona as first author – report the results of experiments that will help to answer these questions (deHaro-Arbona et al., 2023; Figure 1).
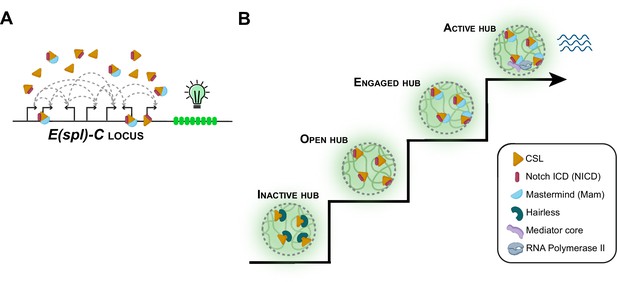
Notch signaling and transcriptional hubs.
(A) The E(spl)-C locus contains multiple genes that are regulated by the Notch signaling pathway. (B) When there is no Notch signal, the transcriptional hub near the E(spl)-C locus is inactive (bottom left): an inactive hub represses gene expression because CSL is in complex with the suppressor protein Hairless. When there is a Notch signal, CSL associates with the Notch intracellular domain (NICD) to create an open hub, which targets Notch-activated genes, and the CSL-NICD complex recruits the co-activator Mastermind to create an engaged hub. A subset of engaged hubs become active (top right) by recruiting a Mediator complex and RNA polymerase II to engage in transcription.
Image credits: Figure created using elements from Biorender.com.
The team focused on the Notch signaling pathway, which becomes activated when a Notch receptor at the plasma membrane of a cell binds to its ligand. This triggers the release of the Notch intracellular domain (NICD) into the cell where it recruits two other proteins: a transcription factor known as CSL, and the co-activator protein Mastermind. Together, with RNA Polymerase II (the protein complex that transcribes DNA) and Mediator (a complex that is also involved in transcription), they regulate the expression of many genes.
To study this process, deHaro-Arbona et al. performed ex vivo imaging of Drosophila larval salivary glands. The experiments looked at a gene locus called E(spl)-C (short for the Enhancer of split Complex), which contains multiple genes that are regulated by Notch signaling. This locus and its regulators (CSL, Mastermind, RNA Polymerase II and Mediator) were each fluorescently labelled and monitored in live cells.
The findings show that Mastermind and CSL form a hub at the E(spl)-C locus when Notch signaling is activated. Intriguingly, the amount of CSL recruited to the hub did not correlate with the number of CSL binding sites at the locus, suggesting that CSL proteins do not interact with these regions in a one-to-one ratio. This more complex type of co-operation, known as non-stoichiometric binding, may be mediated by weak protein-protein interactions. As intrinsically disordered regions in proteins are often implicated in such interactions (Chong et al., 2018), deHaro-Arbona et al. investigated the role of these regions in NICD, CSL and Mastermind. They found that while the disordered region of NICD targeted the E(spl)-C locus, the disordered regions in CSL and Mastermind made only minor contributions to the hub. It appears, therefore, that the hub only partially relies on intrinsically disordered regions.
This is consistent with prior observations that not all hubs are equal and, instead, they consist of local microenvironments of various sizes, compositions and biophysical properties, which are generally dynamic and evolve during transcription (Sharp et al., 2022). Hubs are generally located in places where molecules rely on slower diffusion kinetics to search for their target (Lu and Lionnet, 2021). This is also the case for Notch activator complexes, which exhibited slow diffusion and a long residence time at E(spl)-C.
To examine how the function of the hub depends on its individual components, deHaro-Arbona et al. inhibited the recruitment of the Mastermind protein. This did not impact the recruitment of CSL to the hub. It also did not prevent Notch signalling from increasing chromatin accessibility, suggesting the hub does not need Mastermind in order to access chromatin. However, another protein in the hub, a Mediator complex named Med13, was not recruited. These findings indicate that while some changes induced by Notch signaling can occur independently of Mastermind, it is essential for recruitment of Med13.
Next, deHaro-Arbona et al. investigated how the Notch hubs impact transcription. Live imaging showed that only a third of hubs recruited the Mediator complex and RNA Polymerase II (which enable transcription), and only a third of cells showed active transcription following signaling. This is consistent with the view that even in the presence of all the necessary transcription factors, active transcription is probabilistic (Lu and Lionnet, 2021).
It remains an open question how promoters decode transcriptional hubs to trigger a specific choreography of RNA Polymerase II activation. A transcriptional hub being present does not always result in bursts of transcriptional activity (Dufourt et al., 2018; Mir et al., 2018). In some contexts, a high local density of newly synthesized transcripts can dissolve hubs (Cho and O’Farrell, 2022; Sharp et al., 2022), allowing hubs to self-limit their existence using RNA-mediated feedback mechanisms. The transient nature of hubs could be contributing to the stochasticity of transcription, but this hypothesis warrants more investigation.
Finally, deHaro-Arbona et al. asked if the action of a Notch transcription hub could persist once cells are no longer subject to a Notch input signal. Optogenetics experiments revealed that loci with prior exposure to Notch signaling were re-activated by subsequent Notch signaling more rapidly than naïve cells, thus displaying a form of ‘memory’. deHaro-Arbona et al. propose this memory might arise from the transcription factor CSL ‘book-marking’ the E(spl)-C locus during mitosis in order to speed up transcriptional reactivation, as has been observed in experiments with other transcription factors in vivo (Bellec et al., 2022; Gonzalez et al., 2021). This is clearly a topic for further research.
By revealing the dynamic nature of these transcriptional hubs (Figure 1), the work of deHaro-Arbona et al. leads to a number of questions. How long do hubs take to form at the target locus after signal induction, and how long do they take to dissolve once the signal stops? How does the hub evolve once transcription is activated, and how might that impact the timing and variability of transcriptional activity? Overall, the work enriches our understanding of hub formation and the role of hubs in modulating transcription, and provides a flexible platform to explore the function of transcriptional hubs in living organisms.
References
-
The control of transcriptional memory by stable mitotic bookmarkingNature Communications 13:1176.https://doi.org/10.1038/s41467-022-28855-y
-
Mitotic memories of gene activityCurrent Opinion in Cell Biology 69:41–47.https://doi.org/10.1016/j.ceb.2020.12.009
-
Transcription factor dynamicsCold Spring Harbor Perspectives in Biology 13:a040949.https://doi.org/10.1101/cshperspect.a040949
Article and author information
Author details
Publication history
Copyright
© 2023, Pimmett and Lagha
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 1,265
- views
-
- 104
- downloads
-
- 0
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Download links
Downloads (link to download the article as PDF)
Open citations (links to open the citations from this article in various online reference manager services)
Cite this article (links to download the citations from this article in formats compatible with various reference manager tools)
Further reading
-
- Chromosomes and Gene Expression
- Genetics and Genomics
Among the major classes of RNAs in the cell, tRNAs remain the most difficult to characterize via deep sequencing approaches, as tRNA structure and nucleotide modifications can each interfere with cDNA synthesis by commonly-used reverse transcriptases (RTs). Here, we benchmark a recently-developed RNA cloning protocol, termed Ordered Two-Template Relay (OTTR), to characterize intact tRNAs and tRNA fragments in budding yeast and in mouse tissues. We show that OTTR successfully captures both full-length tRNAs and tRNA fragments in budding yeast and in mouse reproductive tissues without any prior enzymatic treatment, and that tRNA cloning efficiency can be further enhanced via AlkB-mediated demethylation of modified nucleotides. As with other recent tRNA cloning protocols, we find that a subset of nucleotide modifications leave misincorporation signatures in OTTR datasets, enabling their detection without any additional protocol steps. Focusing on tRNA cleavage products, we compare OTTR with several standard small RNA-Seq protocols, finding that OTTR provides the most accurate picture of tRNA fragment levels by comparison to "ground truth" Northern blots. Applying this protocol to mature mouse spermatozoa, our data dramatically alter our understanding of the small RNA cargo of mature mammalian sperm, revealing a far more complex population of tRNA fragments - including both 5′ and 3′ tRNA halves derived from the majority of tRNAs – than previously appreciated. Taken together, our data confirm the superior performance of OTTR to commercial protocols in analysis of tRNA fragments, and force a reappraisal of potential epigenetic functions of the sperm small RNA payload.
-
- Chromosomes and Gene Expression
- Genetics and Genomics
Models of nuclear genome organization often propose a binary division into active versus inactive compartments yet typically overlook nuclear bodies. Here, we integrated analysis of sequencing and image-based data to compare genome organization in four human cell types relative to three different nuclear locales: the nuclear lamina, nuclear speckles, and nucleoli. Although gene expression correlates mostly with nuclear speckle proximity, DNA replication timing correlates with proximity to multiple nuclear locales. Speckle attachment regions emerge as DNA replication initiation zones whose replication timing and gene composition vary with their attachment frequency. Most facultative LADs retain a partially repressed state as iLADs, despite their positioning in the nuclear interior. Knock out of two lamina proteins, Lamin A and LBR, causes a shift of H3K9me3-enriched LADs from lamina to nucleolus, and a reciprocal relocation of H3K27me3-enriched partially repressed iLADs from nucleolus to lamina. Thus, these partially repressed iLADs appear to compete with LADs for nuclear lamina attachment with consequences for replication timing. The nuclear organization in adherent cells is polarized with nuclear bodies and genomic regions segregating both radially and relative to the equatorial plane. Together, our results underscore the importance of considering genome organization relative to nuclear locales for a more complete understanding of the spatial and functional organization of the human genome.






