Viral Variants: How mutations affect function
The COVID-19 pandemic has inflicted enormous societal, economic, and personal costs to populations around the world. It has resulted in hundreds of millions of infections, millions of deaths as well as lingering effects such as persistent symptoms and long COVID. Combatting the virus that causes COVID-19, known as SARS-CoV-2, as well as other viruses that could cause future pandemics, requires an understanding of the basic mechanisms through which viruses evolve and exert their effects.
Every person infected with COVID-19 carries their own pool of viral variants, which each contain a unique set of mutations in their genome. State-of-the-art sequencing technology has allowed an organization known as the Global Initiative on Sharing All Influenza Data to create a SARS-CoV-2 repository, which currently contains around 17 million different viral genome sequences (Elbe and Buckland-Merrett, 2017). This provides unparalleled insight into how mutations correlate with the evolution and emergence of SARS-CoV-2 variants of concern, such as delta and omicron.
However, despite this wealth of available sequencing information, our understanding of how these genetic changes actually impact how variants behave, such as their ability to infect cells and the severity of symptoms they cause, is lacking. Now, in eLife, Peter Schuck and colleagues at the National Institutes of Health – including Ai Nguyen as first author – report how mutations affect a structural protein in SARS-CoV-2 that is responsible for packaging and protecting the viral genome (Nguyen et al., 2024).
Nguyen et al. focused their study on nucleocapsid protein, the most abundant SARS-CoV-2 protein in infected cells, which is typically the molecule probed by at-home tests for the virus (Finkel et al., 2021; Frank et al., 2022). The primary role of the nucleocapsid protein is to package the genomic material of SARS-CoV-2 into the shell of the viral particle. But nucleocapsid protein also binds to many proteins in the host cell, which can modify stress and immune responses that may kill the virus or affect the severity of the infection (Chen et al., 2020; Gordon et al., 2020; Li et al., 2020). These interactions are determined by the structure and biophysical properties of nucleocapsid protein, such as its electrical charge and hydrophobicity.
By studying available genetic sequences of SARS-CoV-2 nucleocapsid proteins, Nguyen et al. found that most mutations lie in intrinsically disordered regions of the protein (Figure 1). These parts of the protein are highly flexible and do not fold into any rigidly defined structure. As such, they are more tolerant to a wider diversity of changes and mutations than other, more highly ordered segments of the protein.
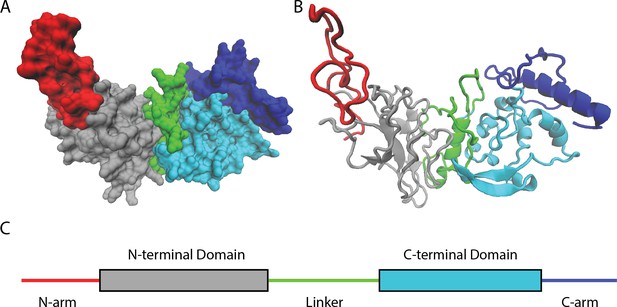
Naturally occurring mutations in the nucleocapsid protein of SARS-CoV-2.
(A) Space filling, (B) ribbon, and (C) domain representation of a single SARS-CoV-2 nucleocapsid protein. Nguyen et al. identified that most mutations occur in intrinsically disordered regions of the protein – the N-arm (red), linker (green), and C-arm (blue) – as opposed to more structured regions, such as the N-terminal (grey) and C-terminal (cyan) domains.
Image credit: Atomic coordinates used from Casasanta et al., 2023 and graphics in (A) and (B) made using Visual Molecular Dynamics (Humphrey et al., 1996).
Nguyen et al. then used computational methods to calculate how the behaviour of nucleocapsid proteins would be expected to change in response to each of the mutations they had detected. This showed that while single mutations in the intrinsically disordered regions cause a broader distribution of effects than those in structured regions of the protein, the biophysical properties of the protein were, surprisingly, largely conserved. Moreover, Nguyen et al. demonstrated that the intrinsically disordered regions show little sequence similarity to related viruses, such as SARS-CoV-1, Middle Eastern Respiratory Syndrome and other coronaviruses. However, the biophysical properties of these regions, such as their polarity and hydrophobicity, were similar across the different viruses.
To investigate further, Nguyen et al. carried out experiments on nucleocapsid proteins which had been purified from SARS-CoV-2 and contained mutations found in several variants of interest. This revealed that when multiple mutations are present, this leads to significant – and sometimes compensatory – biophysical changes. This included changes in the thermal stability of the protein, its ability to self-associate into multi-protein complexes, and how it assembles into larger structures. These effects demonstrate how mutations in intrinsically disordered regions can impact the entire structure and function of a protein. Additionally, this emphasizes why it is difficult to assign variant properties and fitness effects to a single mutation.
The findings of Nguyen et al. suggest that the biophysical properties of intrinsically disordered regions – such as their charge, polarity and hydrophobicity – may contribute to determining the behavior and interactions of nucleocapsid proteins. This could also be the case for other SARS-CoV-2 proteins, as well as proteins in other infectious viruses. These properties may place constraints on the mutations that nucleocapsid proteins can tolerate while still remaining functional. Furthermore, the findings of Nguyen et al. could provide a foundation for understanding how genomic sequencing data can be used to predict the effects of mutations in viruses.
References
-
Structural insights of the SARS-CoV-2 nucleocapsid protein: Implications for the inner-workings of rapid antigen testsMicroscopy and Microanalysis 29:649–657.https://doi.org/10.1093/micmic/ozac036
-
VMD: visual molecular dynamicsJournal of Molecular Graphics 14:33–38.https://doi.org/10.1016/0263-7855(96)00018-5
Article and author information
Author details
Publication history
Copyright
© 2024, Kuhlman
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 672
- views
-
- 79
- downloads
-
- 0
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Download links
Downloads (link to download the article as PDF)
Open citations (links to open the citations from this article in various online reference manager services)
Cite this article (links to download the citations from this article in formats compatible with various reference manager tools)
Further reading
-
- Immunology and Inflammation
- Microbiology and Infectious Disease
Pseudomonas aeruginosa (PA) is an opportunistic, frequently multidrug-resistant pathogen that can cause severe infections in hospitalized patients. Antibodies against the PA virulence factor, PcrV, protect from death and disease in a variety of animal models. However, clinical trials of PcrV-binding antibody-based products have thus far failed to demonstrate benefit. Prior candidates were derivations of antibodies identified using protein-immunized animal systems and required extensive engineering to optimize binding and/or reduce immunogenicity. Of note, PA infections are common in people with cystic fibrosis (pwCF), who are generally believed to mount normal adaptive immune responses. Here, we utilized a tetramer reagent to detect and isolate PcrV-specific B cells in pwCF and, via single-cell sorting and paired-chain sequencing, identified the B cell receptor (BCR) variable region sequences that confer PcrV-specificity. We derived multiple high affinity anti-PcrV monoclonal antibodies (mAbs) from PcrV-specific B cells across three donors, including mAbs that exhibit potent anti-PA activity in a murine pneumonia model. This robust strategy for mAb discovery expands what is known about PA-specific B cells in pwCF and yields novel mAbs with potential for future clinical use.
-
- Microbiology and Infectious Disease
The persistence of latent viral reservoirs remains the major obstacle to eradicating human immunodeficiency virus (HIV). We herein found that ICP34.5 can act as an antagonistic factor for the reactivation of HIV latency by herpes simplex virus type I (HSV-1), and thus recombinant HSV-1 with ICP34.5 deletion could more effectively reactivate HIV latency than its wild-type counterpart. Mechanistically, HSV-ΔICP34.5 promoted the phosphorylation of HSF1 by decreasing the recruitment of protein phosphatase 1 (PP1α), thus effectively binding to the HIV LTR to reactivate the latent reservoirs. In addition, HSV-ΔICP34.5 enhanced the phosphorylation of IKKα/β through the degradation of IκBα, leading to p65 accumulation in the nucleus to elicit NF-κB pathway-dependent reactivation of HIV latency. Then, we constructed the recombinant HSV-ΔICP34.5 expressing simian immunodeficiency virus (SIV) env, gag, or the fusion antigen sPD1-SIVgag as a therapeutic vaccine, aiming to achieve a functional cure by simultaneously reactivating viral latency and eliciting antigen-specific immune responses. Results showed that these constructs effectively elicited SIV-specific immune responses, reactivated SIV latency, and delayed viral rebound after the interruption of antiretroviral therapy (ART) in chronically SIV-infected rhesus macaques. Collectively, these findings provide insights into the rational design of HSV-vectored therapeutic strategies for pursuing an HIV functional cure.






