Vaccines: How do adjuvants enhance immune responses?
The process by which a vaccine enhances immunity against a disease involves a wide range of cell types. It starts with cells called antigen presenting cells (APCs) internalizing and processing antigens from the vaccine. These APCs then present the antigens on MHC II molecules at the cell surface, a process that can activate cells called T cells. T cell activation is a prerequisite for other immune cells called B cells to produce the antibodies that are crucial to the immune response.
Adjuvants are substances added to vaccines that enhance the immune response to antigens and ultimately, improve immunity. It is known that some, such as MPLA and CpG, directly activate proteins found on APCs called pattern recognition receptors (Reed et al., 2013; Didierlaurent et al., 2014). Yet how adjuvants influence the magnitude and quality of adaptive immune responses through APCs remains unclear. It was recently shown that adding an adjuvant to a vaccine may influence the specific region of a vaccine antigen that an antibody recognizes (Maeda et al., 2017; Chung et al., 2015). This suggests that CD4+ T cells, required for fine-tuning B cell antibody responses, are also impacted by adjuvants.
Now, in eLife, Bin Li, Peng Wang, Chao Wu of The Eighth Affiliated Hospital of Sun Yat-sen University and colleagues – including Jin Zhang as joint first author with Li – report the results of experiments which shed light on how adjuvants work (Li et al., 2024). The experiments involved administering a vaccine containing a protein antigen from the bacteria H. pylori to mice: in some cases, the vaccine also contained MPLA or CpG as an adjuvant, and in other cases it did not. Interestingly, the various adjuvanted or unadjuvanted vaccination conditions differently impacted which protein sites were most reactive with T cells. When APCs were exposed to H. pylori proteins in the presence of these adjuvants, peptide antigens with low affinity for MHC II were more likely to be presented, and the number of different presented peptide sequences was low. In contrast, when the vaccine did not contain adjuvants, peptide antigens with high affinity for MHC II were more likely to be presented, and the number of different presented sequences was higher (Figure 1).
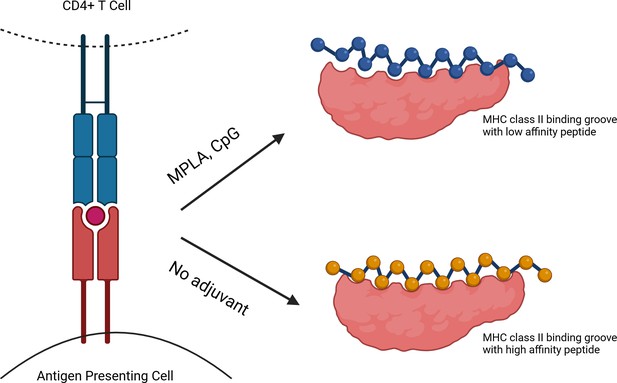
The impact of adjuvants on interactions between APCs and peptide antigens.
A CD4+ T cell (top left) may be activated when the T cell receptor (blue) interacts with an MHC II molecule (red) and a peptide antigen (red circle) on the surface of an antigen presenting cell (APC; bottom left). APCs exposed to adjuvants (such as MPLA or CpG; top right) in the presence of vaccine antigens present peptide antigens (blue) that have lower affinity molecular interactions with MHC II (pink), and present fewer different peptides. In contrast, in the absence of adjuvants (bottom right), vaccine peptides with higher affinity molecular interactions (yellow) with MHC II are presented, as well as a broader peptide repertoire.
© 2024, BioRender Inc. Figure 1 was created using BioRender, and is published under a CC BY-NC-ND license. Further reproductions must adhere to the terms of this license.
When a peptide with low affinity for MHC II was used to vaccinate mice, it was found that significantly less peptide was needed to activate CD4+ T cells compared to responses elicited by vaccination with high-affinity peptides. Taken together, these findings support previous observations that T cell responses to a protein are often focused on a narrow array of peptide antigens (Sant et al., 2005; Baumgartner and Malherbe, 2010), but they also show that different adjuvants have different effects on how T cells respond to peptide antigens. It is an important distinction that adjuvants, beyond increasing the magnitude of a given immune response, may also fine-tune the portion of the antigen targeted by T cells.
How peptide antigens with low affinity for MHC II and a narrow antigen repertoire might impact the CD4+ T cell response remains an open question. Some data suggest that the threshold for activating T cells is influenced by adjuvants that can activate pattern recognition receptors, and that the complexes formed by low-affinity peptides and MHC II molecules provide stronger signals to T cell receptors, promoting their activation (Malherbe et al., 2008; Baumgartner et al., 2010).
Alternatively, it has also been shown that a narrow peptide repertoire can enhance T cell responses (Santambrogio, 2022). Could it be that restricting the number of antigens that APCs present (by increasing selection of low-affinity peptides) narrows the peptide repertoire and therefore focuses the T cell response? Li et al. were able to demonstrate that this phenomenon of peptide selection was not due to differences in which antigens were initially taken up by APCs, suggesting that these adjuvants influence antigen processing. Exactly which steps of antigen processing are affected remains to be explored. Might these steps be impacted by pattern recognition receptor signaling? Additionally, it remains to be explored whether these mechanisms are at play in the human immune system, which has more complex MHC II structures than those found in mice.
The findings of Li et al. bring us closer to understanding how adjuvants that activate pattern recognition receptors actually work. By influencing antigen processing and focusing peptide presentation by APCs, adjuvants change the nature of the CD4+ T cell response to a vaccine. This underscores how finely and specifically these adjuvants might influence a vaccine response. Modeling how a given adjuvant fine-tunes the MHC II peptide repertoire may be an important early step in vaccine development. Tackling the development of more challenging vaccines – or optimizing the use of vaccines in populations across the age spectrum – may ultimately rest on our ability to understand and take advantage of the precise mechanisms of these types of adjuvants.
References
-
Peptide-MHC class II complex stability governs CD4 T cell clonal selectionJournal of Immunology 184:573–581.https://doi.org/10.4049/jimmunol.0902107
-
Key roles of adjuvants in modern vaccinesNature Medicine 19:1597–1608.https://doi.org/10.1038/nm.3409
-
Molecular determinants regulating the plasticity of the MHC Class II immunopeptidomeFrontiers in Immunology 13:878271.https://doi.org/10.3389/fimmu.2022.878271
Article and author information
Author details
Publication history
Copyright
© 2024, Rapaka
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 639
- views
-
- 83
- downloads
-
- 0
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Download links
Downloads (link to download the article as PDF)
Open citations (links to open the citations from this article in various online reference manager services)
Cite this article (links to download the citations from this article in formats compatible with various reference manager tools)
Further reading
-
- Cell Biology
- Immunology and Inflammation
The endothelial blood-brain barrier (BBB) strictly controls immune cell trafficking into the central nervous system (CNS). In neuroinflammatory diseases such as multiple sclerosis, this tight control is, however, disturbed, leading to immune cell infiltration into the CNS. The development of in vitro models of the BBB combined with microfluidic devices has advanced our understanding of the cellular and molecular mechanisms mediating the multistep T-cell extravasation across the BBB. A major bottleneck of these in vitro studies is the absence of a robust and automated pipeline suitable for analyzing and quantifying the sequential interaction steps of different immune cell subsets with the BBB under physiological flow in vitro. Here, we present the under-flow migration tracker (UFMTrack) framework for studying immune cell interactions with endothelial monolayers under physiological flow. We then showcase a pipeline built based on it to study the entire multistep extravasation cascade of immune cells across brain microvascular endothelial cells under physiological flow in vitro. UFMTrack achieves 90% track reconstruction efficiency and allows for scaling due to the reduction of the analysis cost and by eliminating experimenter bias. This allowed for an in-depth analysis of all behavioral regimes involved in the multistep immune cell extravasation cascade. The study summarizes how UFMTrack can be employed to delineate the interactions of CD4+ and CD8+ T cells with the BBB under physiological flow. We also demonstrate its applicability to the other BBB models, showcasing broader applicability of the developed framework to a range of immune cell-endothelial monolayer interaction studies. The UFMTrack framework along with the generated datasets is publicly available in the corresponding repositories.
-
- Immunology and Inflammation
The gut biome, a complex ecosystem of micro- and macro-organisms, plays a crucial role in human health. A disruption in this evolutive balance, particularly during early life, can lead to immune dysregulation and inflammatory disorders. ‘Biome repletion’ has emerged as a potential therapeutic approach, introducing live microbes or helminth-derived products to restore immune balance. While helminth therapy has shown some promise, significant challenges remain in optimizing clinical trials. Factors such as patient genetics, disease status, helminth species, and the optimal timing and dosage of their products or metabolites must be carefully considered to train the immune system effectively. We aim to discuss how helminths and their products induce trained immunity as prospective to treat inflammatory and autoimmune diseases. The molecular repertoire of helminth excretory/secretory products (ESPs), which includes proteins, peptides, lipids, and RNA-carrying extracellular vesicles (EVs), underscores their potential to modulate innate immune cells and hematopoietic stem cell precursors. Mimicking natural delivery mechanisms like synthetic exosomes could revolutionize EV-based therapies and optimizing production and delivery of ESP will be crucial for their translation into clinical applications. By deciphering and harnessing helminth-derived products’ diverse modes of action, we can unleash their full therapeutic potential and pave the way for innovative treatments.






